WRF-LES installation and use on the server

1. Installation
1.1 Script
Environment:
- OS: Ubuntu 22.04.5 LTS without sudo permission
- Compilers Version: gcc 11.4.0 (system native)
Before installing the libraries, these paths need to be set by adding the following sentence at the end of bashrc. Please note the DIR needs to be replaced with the location on your computer.
Installation of LIBRARY like netcdf, zlib, libpng… and compilation of WRF
if you use this bash for the first time, it’s better to run the script line by line
#!/bin/bash
## WRF installation with parallel process.
# Download and install required library for WRF.
# The WRF folder will be under HOME
# Zhipeng Pei <zhipeng.pei@whu.edu.cn>
# Tested in Ubuntu 22.04.5 LTS
# Ctrl+D to comment and Ctrl+shift+D to uncomment line in KATE
## Directory Listing
export HOME=`cd;pwd`
mkdir -p $HOME/WRF
cd $HOME/WRF
mkdir -p Downloads
mkdir -p Library
## Downloading Libraries
cd Downloads
wget -c https://support.hdfgroup.org/ftp/HDF5/releases/hdf5-1.10/hdf5-1.10.5/src/hdf5-1.10.5.tar.gz && echo "hdf5 download done"
wget -c https://downloads.unidata.ucar.edu/netcdf-c/4.9.0/netcdf-c-4.9.0.tar.gz
wget -c https://downloads.unidata.ucar.edu/netcdf-fortran/4.6.0/netcdf-fortran-4.6.0.tar.gz
wget -c https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/jasper-1.900.1.tar.gz
wget -c https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/libpng-1.2.50.tar.gz
wget -c https://github.com/madler/zlib/releases/download/v1.2.13/zlib-1.2.13.tar.gz
wget -c https://download.open-mpi.org/release/open-mpi/v4.1/openmpi-4.1.6.tar.gz
wget -c https://www2.mmm.ucar.edu/wrf/OnLineTutorial/compile_tutorial/tar_files/mpich-3.0.4.tar.gz
echo "All download done"
# Compilers
export DIR=$HOME/WRF/Library
export CC=gcc
export CXX=g++
export FC=gfortran
export F77=gfortran
export JASPERLIB=$DIR/grib2/lib
export JASPERINC=$DIR/grib2/include
export LDFLAGS=-L$DIR/grib2/lib
export CPPFLAGS=-I$DIR/grib2/include
export fallow_argument=-fallow-argument-mismatch
export boz_argument=-fallow-invalid-boz
export FFLAGS="$fallow_argument $boz_argument -m64"
export FCFLAGS="$fallow_argument $boz_argument -m64"
# ## Install NETCDF C Library
# # --disable-netcdf-4 --disable-shared is necessary
cd $HOME/WRF/Downloads
tar -xvzf netcdf-c-4.9.0.tar.gz
cd netcdf-c-4.9.0/
make clean
./configure --prefix=$DIR/netcdf --disable-dap --disable-netcdf-4 --disable-shared
make check
make install
export PATH=$DIR/netcdf/bin:$PATH
export NETCDF=$DIR/netcdf
# ## NetCDF fortran library
export LIBS="-lnetcdf -lz"
cd $HOME/WRF/Downloads
tar -xvzf netcdf-fortran-4.6.0.tar.gz
cd netcdf-fortran-4.6.0/
make clean
./configure --prefix=$DIR/netcdf --disable-dap --disable-netcdf-4 --disable-shared
make check
make install
export PATH=$DIR/netcdf/bin:$PATH
export NETCDF=$DIR/netcdf
## openmpi, it will take a lot of time
cd $HOME/WRF/Downloads
tar -xzf openmpi-4.1.6.tar.gz
cd openmpi-4.1.6
make clean
./configure --prefix=$DIR/openmpi
make
make install
export PATH=$DIR/openmpi/bin:$PATH
## MPICH there are some problem in make for my compilation
# cd $HOME/WRF/Downloads
# tar xzvf mpich-3.0.4.tar.gz #or just .tar if no .gz present
# cd mpich-3.0.4
# ./configure --prefix=$DIR/mpich
# make
# make install
# export PATH=$DIR/mpich/bin:$PATH
# zlib
cd $HOME/WRF/Downloads
tar -xvzf zlib-1.2.13.tar.gz
cd zlib-1.2.13/
make clean
./configure --prefix=$DIR/grib2
make
make install
# # hdf5 library for netcdf4 functionality
# # make check will take a lot of time
# cd $HOME/WRF/Downloads
# tar -xvzf hdf5-1.10.5.tar.gz
# cd hdf5-1.10.5
# make clean
# ./configure --prefix=$DIR --with-zlib=$DIR --enable-hl --enable-fortran
# make check
# make install
#
# export HDF5=$DIR
# export LD_LIBRARY_PATH=$DIR/lib:$LD_LIBRARY_PATH
# # libpng
cd $HOME/WRF/Downloads
tar -xvzf libpng-1.2.50.tar.gz
cd libpng-1.2.50/
make clean
./configure --prefix=$DIR/grib2
make
make install
# # JasPer
cd $HOME/WRF/Downloads
tar -xvzf jasper-1.900.1.tar.gz
cd jasper-1.900.1/
make clean
./configure --prefix=$DIR/grib2
make
make install
## export PATH and LD_LIBRARY_PATH
echo "export PATH=$DIR/bin:$PATH" >> ~/.bashrc
echo "export LD_LIBRARY_PATH=$DIR/lib:$LD_LIBRARY_PATH" >> ~/.bashrc
source ~/.bashrc
# ############################ WRF 4.2.2 #################################
# ## WRF v4.2.2
# ########################################################################
cd $HOME/WRF
git clone https://github.com/wrf-model/WRF
cd WRF
git checkout release-v4.2.2
./clean -a
export NETCDF_classic=1
./configure # select 34 and 1 for gfortran and distributed memory
./compile em_les >& compile_bash_step_by_step.log
Check for errors, there should be nothing on the console. And there should be ideal.exe and wrf.exe under WRF/test/em_les folder
cat -n compile_bash_step_by_step.log | grep Error
If you want to recompile the WRF on the another console, it’s recommanded add these lines to the bash_profile and source it because the variable (like NETCDF) is temporary
$ gedit ~/.bash_profile
export HOME=`cd;pwd`
export DIR=$HOME/WRF/Library
export CC=gcc
export CXX=g++
export FC=gfortran
export F77=gfortran
export JASPERLIB=$DIR/grib2/lib
export JASPERINC=$DIR/grib2/include
export LDFLAGS=-L$DIR/grib2/lib
export CPPFLAGS=-I$DIR/grib2/include
export fallow_argument=-fallow-argument-mismatch
export boz_argument=-fallow-invalid-boz
export FFLAGS="$fallow_argument $boz_argument -m64"
export FCFLAGS="$fallow_argument $boz_argument -m64"
export NETCDF=$DIR/netcdf
export NETCDF_classic=1
$ source ~/.bash_profile
1.2 Problems
Some of the problems I’ve had
Problem 1:
more undefined references to `__module_optional_input_MOD_st_input' follow
collect2: error: ld returned 1 exit status
As WRF forum decribed, Using openmpi not MPICH for Ubuntu 22.04. If you have sudo permission, just
sudo apt install libopenmpi-dev libhdf5-openmpi-dev
If you don’t have sudo permission, install openmpi manually as described in my bash file above.
Check mpirun, and it should be
$ mpirun --version
mpirun (Open MPI) 4.1.6
Report bugs to http://www.open-mpi.org/community/help/
Problem 2:
************************** W A R N I N G ************************************
NETCDF4 IO features are requested, but this installation of NetCDF
/home/zppei/WRF/Library/netcdf
DOES NOT support these IO features.
Please make sure NETCDF version is 4.1.3 or later and was built with
--enable-netcdf4
OR set NETCDF_classic variable
bash/ksh : export NETCDF_classic=1
echo csh : setenv NETCDF_classic 1
Then re-run this configure script
!!! configure.wrf has been REMOVED !!!
*****************************************************************************
Like the official wrf guide, I didn’t install hdf5, although you may read in other blogs that hdf installation is important for netcdf installation. I asked ChatGPT and get the answer about hdf5 and netcdf.
In summary, the HDF5 (Hierarchical Data Format 5) library is necessary for NetCDF-4. But we can disable NetCDF-4 support without HDF5 ./configure --disable-netcdf-4. That’s the reason why I get Problem 2. Take easy, just the following command will solve the problem.
export NETCDF_classic=1
Problem 3:
$ cat -n compile_bash_step_by_step.log | grep Error
165 Error: Dummy argument ?datasetname? with INTENT(IN) in variable definition context (actual argument to INTENT = OUT/INOUT) at (1)
173 Error: Rank mismatch between actual argument at (1) and actual argument at (2) (rank-1 and scalar)
178 Error: Type mismatch in argument ?field? at (1); passed INTEGER(4) to REAL(8)
davegill suggested use new release of WRF.
git clone https://github.com/wrf-model/WRF
cd WRF
git checkout release-v4.2.2
2. Running ideal LES
2.1 namelist.input
Here is an example of my namelist.input
2.1.1 Description
Here is the description of namelist.input from ucar.
2.1.2 Nested domain

2.1.3 Output variables and frequency
According to the UCAR website
- The first step is to create a text file (e.g., my_file_d0X.txt), for each domain, and define the name of that file in the &time_control section of namelist.input, as indicated below.
&time_control
iofields_filename = "myoutfields_d01.txt", "myoutfields_d02.txt","myoutfields_d03.txt" ! Name of file that tells WRF what to output
ignore_iofields_warning = .true.
- Contents of the text file associate a stream ID (0 is the default history and input) with a variable, and whether the field is added or removed. Following are a few examples.
-:h:0:RAINC,RAINNC
removes the fields RAINC and RAINNC from the standard history file.
+:h:7:RAINC,RAINNC
adds the fields RAINC and RAINNC to an output stream #7, which would create a separate file from the wrfout* files.
Available options are
- + or -, add or remove a variable
- 0-24, which stream (integer)
- i or h, input or history
- field name in the Registry – this is the first string in quotes.
If you are interested in outputting variables into a new stream (i.e., not the default history stream 0), then the following namelist variables will also be necessary (example for stream 7):
auxhist7_outname = “yourstreamname_d<domain>_<date>”
auxhist7_interval = 360, 360,
frames_per_auxhist7 = 1, 1,
io_form_auxhist7 = 2
Here is an example from Brian Blaylock
2.1.4 Time_step
According to the README.namelist
time step for integration in integer seconds recommend 6*dx (in km) for typical real-data cases
2.2 input.sounding
Here is an example of my input_sounding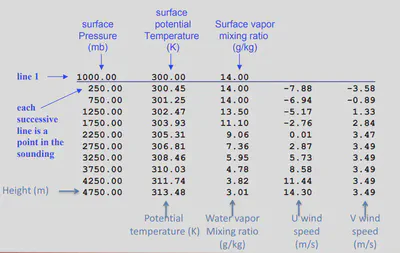
2.3 How to run model with multi processor
mpirun -np 6 ./wrf.exe use 6 logical processors.
The allocation of these 6 processes can occur on one or multiple physical CPUs, depending on how the scheduler distributes them.
To understand the difference between CPUs, Cores and logical processors: For example, each of your physical CPUs has 14 cores. If hyper-threading is enabled, each core can run 2 threads, resulting in 28 logical processors per physical CPU. Many thanks to ChatGPT :).
If you dont know the number of your CPU/cores, use
lscpu
to show the information.
2.4 Parallel
For your smallest-sized domain: ((e_we)/25) * ((e_sn)/25) = most amount of processors you should use
For your largest-sized domain: ((e_we)/100) * ((e_sn)/100) = least amount of processors you should use
2.3 Problems
Problem 1: Strange filenames due to colon : in Mac finder
WRF outputs NetCDF filenames (like auxhist24_d02_0001-01-01_00:00:00) that contain timestamps (often including a colon :), which on Mac finder systems is an illegal character or causes the filename to be garbled.
you can use the mvcommand to rename a file that contains a colon:
for file in wrfout*; do
new_file=$(echo $file | sed 's/:/_/g')
mv "$file" "$new_file"
done